Difference between revisions of "Getting started"
| (45 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | The best way to learn ''Dynamo'' on your own is to follow the materials in one of our 2 day-workshops such as [[PSI Workshop 2023]]. These materials are sorted and described in [[#Suggested order of walkthroughs | the section of suggested walkthroughs]]. | ||
| − | |||
| + | == Starters guide == | ||
| + | Concerning the task of subtomogram averaging, you can get a plunge-freeze immersion in ''Dynamo'' for from scratch, by following the [[Starters guide| starters guide]]. This is a short hands-on complete walkthrough through the main functionalities in ''Dynamo'': [[Viewing tomograms|viewing]] and [[Catalogue|organizing]] tomograms, [[Particle extraction|extracting particles]], [[Subtomogram alignment|aligning]], averaging and [[Classification|classifying]] subtomograms, and [[Adaptive bandpass filtering|avoiding overfitting]]. | ||
| − | + | ||
| + | == Getting help == | ||
| + | ''Dynamo'' offers several ways of [[Getting help|getting help]] on specific subjects.... and of course, you can always [[https://groups.google.com/forum/#!forum/dynamo-for-cryo-electron-tomography contact our forum in Google Groups]] | ||
| + | |||
| + | |||
| + | |||
| + | ==Suggested order of walkthroughs== | ||
| + | |||
| + | The following is a subset of the available walkthroughs that give a general overview of the main modules and tools that are available in ''Dynamo''. | ||
| + | |||
| + | ===Core module of ''Dynamo'': models and STA=== | ||
| + | [[File:orthoPickMain.png|thumb|upright|250px| Clicking particles in the [[Starters guide ]] ]] | ||
| + | The first walkthrough is the [[Starters guide]]. It is used to learn the basic functionalities of ''Dynamo'' for tomogram annotation, particle picking and execution of STA experiments.<br><br> | ||
| + | |||
| + | The following steps are the walkthroughs [[Dipole set models]], [[Walkthrough on filament models]], [[Walkthrough on vesicle models]] and [[Walkthrough on membrane models]]. These walkthroughs introduce how sample architecture can be encoded and exploited in ''Dynamo''.<br><br> | ||
| + | |||
| + | Walkthrough [[Tips for management of tomographic data sets]] gives suggestions on how to manage the data used and generated by ''Dynamo'', giving context on the catalogue object used by ''Dynamo'' to keep track of a dataset.<br><br> | ||
| + | |||
| + | |||
| + | Finally, the walkthrough [[Advanced starters guide]] shows how the previous ideas can be put together in a practical case. | ||
| + | |||
| + | [[ File:FhvTmsliceUpdates.png |thumb|center|600px| Modelling a membrane in the [[Advanced starters guide]]]] | ||
| + | |||
| + | ===Towards a full Cryo-ET processing pipeline=== | ||
| + | |||
| + | The following walkthroughs have been ordered to give a general view of the different ''Dynamo'' functionalities that are useful at different steps of a general Cryo-ET processing pipeline. | ||
| + | |||
| + | |||
| + | [[File:General_pipeline.png|centre|thumb|800px| Cryoem general pipeline shown from grid preparation to high-resolution final average. The steps presented in the figure are, from left to right: grid preparation, data acquisition in the microscope, creation of the tilt series, contrast transfer function extraction, tilt series alignment, CTF correction, tomogram generation and modelling, subtomogram averaging and finally recovery of the high resolution average.]] | ||
| + | |||
| + | |||
| + | |||
| + | ====Tilt series alignment==== | ||
| + | ''Dynamo'' includes a module for tilt series alignment. [[Walkthrough on manual marker clicking]] introduces the main concepts of tilt series alignment in ''Dynamo''.<br><br> | ||
| + | |||
| + | [[Walkthrough on GUI based tilt series alignment]] and then [[Walkthrough on command line based tilt series alignment]] present the ''Dynamo'' functionalities for automatic tilt series alignment using fiducials. | ||
| + | |||
| + | ====Managing multiple tomograms==== | ||
| + | |||
| + | The walkthrough [[Getting a Structure from Multiple Tomograms of HIV Capsids (walkthrough)]] present how to set up efficient processing pipelines to annotate and pick particles in datasets with multiple tomograms. | ||
| + | |||
| + | ====Particle picking with template matching==== | ||
| + | |||
| + | A template matching module is available in ''Dynamo''. [[Walkthrough for template matching]] presents the basic functionalities of the module.<br><br> | ||
| + | |||
| + | [[Walkthrough for GUI-based template matching]] introduces how the functionalities are integrated in ''Dynamo'' GUI. | ||
| + | |||
| + | ====Classification with PCA==== | ||
| + | |||
| + | [[Walkthrough on PCA through the command line]] introduces how ''Dynamo'' allows the users to use PCA to then classify sets of particles. | ||
| + | |||
| + | ====Inspection of ''Dynamo'' results==== | ||
| + | |||
| + | [[Walkthrough on placement of averages on table positions]] describes how to take advantage of different ''Dynamo'' tools to generate representations of the data. | ||
| + | |||
| + | ====Step-by-step guide==== | ||
| + | |||
| + | Finally, the [[High resolution walkthrough with HIV1]] can be done to see how the various functionalities of ''Dynamo'' can be used together in a practical step-by-step walkthrough going from raw microscope data of a cryo-electron tomography experiment to a high-resolution average. | ||
| + | |||
| + | == Set of walkthroughs == | ||
| + | The following table lists a set of suggested walkthroughs for different topics on Cryo-ET data processing using ''Dynamo''. The full list of walkthroughs can be accessed through [[:Category:Walkthroughs | its page]]. | ||
| + | {| class="wikitable" style="margin:auto" | ||
| + | |+ Set of available walkthroughs sorted by topic | ||
| + | |- | ||
| + | ! scope="col" style="background-color:#4EDFD7" |Topic | ||
| + | ! scope="col" style="background-color:#4EDFD7" |Walkthrough | ||
| + | |- | ||
| + | ! scope="row" rowspan="5"| General | ||
| + | | style="background-color:#FFFFE5" | [[Starters guide]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Advanced starters guide]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Tips for management of tomographic data sets]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Getting a Structure from Multiple Tomograms of HIV Capsids (walkthrough)]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[High resolution walkthrough with HIV1]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="4"| Models | ||
| + | | style="background-color:#ffffcc" | [[Dipole set models]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough on filament models]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough on vesicle models]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough on membrane models]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="1" | PCA | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on PCA through the command line]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="2"| STA and MRA | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough basic multireference]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough on adaptive bandpass filtering]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="4"| Tilt series alignment | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on manual marker clicking]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on GUI based tilt series alignment]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on command line based tilt series alignment]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on localized reconstruction]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="4"| Template matching | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough for template matching]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough for GUI-based template matching]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Walkthrough for model-aware template matching]] | ||
| + | |- | ||
| + | | style="background-color:#ffffcc" | [[Programmatic GUI customization for template matching]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="3"| Visualization | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on placement of averages on table positions]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on creation of 3d scenes]] | ||
| + | |- | ||
| + | | style="background-color:#FFFFE5" | [[Walkthrough on hexagonal lattice coherence through neighborhood analysis]] | ||
| + | |- | ||
| + | ! scope="row" rowspan="1" | Others | ||
| + | | style="background-color:#ffffcc" | [[Integration with Warp and M]] | ||
| + | |- | ||
| + | |} | ||
Latest revision as of 14:02, 23 September 2025
The best way to learn Dynamo on your own is to follow the materials in one of our 2 day-workshops such as PSI Workshop 2023. These materials are sorted and described in the section of suggested walkthroughs.
Starters guide
Concerning the task of subtomogram averaging, you can get a plunge-freeze immersion in Dynamo for from scratch, by following the starters guide. This is a short hands-on complete walkthrough through the main functionalities in Dynamo: viewing and organizing tomograms, extracting particles, aligning, averaging and classifying subtomograms, and avoiding overfitting.
Getting help
Dynamo offers several ways of getting help on specific subjects.... and of course, you can always [contact our forum in Google Groups]
Suggested order of walkthroughs
The following is a subset of the available walkthroughs that give a general overview of the main modules and tools that are available in Dynamo.
Core module of Dynamo: models and STA
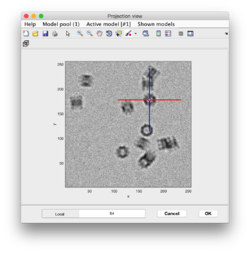
The first walkthrough is the Starters guide. It is used to learn the basic functionalities of Dynamo for tomogram annotation, particle picking and execution of STA experiments.
The following steps are the walkthroughs Dipole set models, Walkthrough on filament models, Walkthrough on vesicle models and Walkthrough on membrane models. These walkthroughs introduce how sample architecture can be encoded and exploited in Dynamo.
Walkthrough Tips for management of tomographic data sets gives suggestions on how to manage the data used and generated by Dynamo, giving context on the catalogue object used by Dynamo to keep track of a dataset.
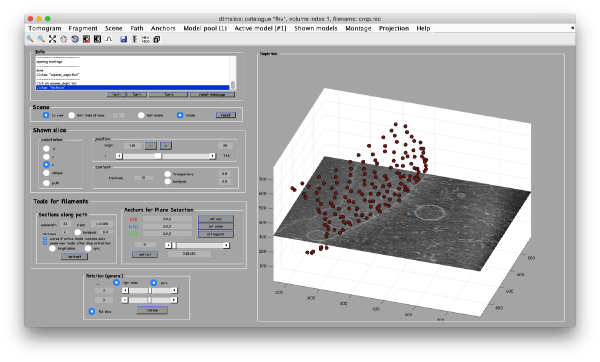
Finally, the walkthrough Advanced starters guide shows how the previous ideas can be put together in a practical case.
Towards a full Cryo-ET processing pipeline
The following walkthroughs have been ordered to give a general view of the different Dynamo functionalities that are useful at different steps of a general Cryo-ET processing pipeline.

Tilt series alignment
Dynamo includes a module for tilt series alignment. Walkthrough on manual marker clicking introduces the main concepts of tilt series alignment in Dynamo.
Walkthrough on GUI based tilt series alignment and then Walkthrough on command line based tilt series alignment present the Dynamo functionalities for automatic tilt series alignment using fiducials.
Managing multiple tomograms
The walkthrough Getting a Structure from Multiple Tomograms of HIV Capsids (walkthrough) present how to set up efficient processing pipelines to annotate and pick particles in datasets with multiple tomograms.
Particle picking with template matching
A template matching module is available in Dynamo. Walkthrough for template matching presents the basic functionalities of the module.
Walkthrough for GUI-based template matching introduces how the functionalities are integrated in Dynamo GUI.
Classification with PCA
Walkthrough on PCA through the command line introduces how Dynamo allows the users to use PCA to then classify sets of particles.
Inspection of Dynamo results
Walkthrough on placement of averages on table positions describes how to take advantage of different Dynamo tools to generate representations of the data.
Step-by-step guide
Finally, the High resolution walkthrough with HIV1 can be done to see how the various functionalities of Dynamo can be used together in a practical step-by-step walkthrough going from raw microscope data of a cryo-electron tomography experiment to a high-resolution average.
Set of walkthroughs
The following table lists a set of suggested walkthroughs for different topics on Cryo-ET data processing using Dynamo. The full list of walkthroughs can be accessed through its page.