I2PC – Instruct Course on Electron Tomography and Subtomogram Averaging 2025
Contents
- 1 The course
- 2 Installation of Dynamo and Matlab
- 3 Computing resources
- 4 Time schedule
- 5 Instructors
The course
The venue
This page describes the contents of the Dynamo-specific part of the course "I2PC – Instruct Course on Electron Tomography and Subtomogram Averaging 2025", which will take part at the premises of the Centro Nacional de Biotecnología in Madrid, from the 9th to the 12th of December 2025.
Contents
Whole pipelines for cryo-ET processing will be covered in the Scipion-related area of the course (days 1 and 2). The Dynamo part will covered selected parts of the pipeline.
Installation of Dynamo and Matlab
Dynamo on your own laptop
To follow the course in your own laptop, please install the Dynamo for Matlab version as explain in Downloads page.
Computing resources
Laptops
Participants can use their own laptops to complete the Dynamo part of the course. For this, participants need to have and active Matlab installation and an Dynamo installation.
- For Matlab, we advise the (R2025b version). If you don't have a license through your home institution, a licence free trial version is available from the Mathworks company.
- The Dynamo Download link is currently down, we will update it later today (17:00 ECT): prior to installation make sure you have a compiler. On Linux you probably have one by default. On Mac, Xcode needs to be installed. On Windows, the recommended option is to install it via Matlab:
https://es.mathworks.com/help/matlab/matlab_external/install-mingw-support-package.html
Dynamo is delivered as a single .tar file. To install it, move the file to the location where you want to install Dynamo, then open a Matlab session and type after the Matlab prompt:
>> tar -xvf dynamo_temp_1.1.579.tar
to untar the installation file. Dynamo will be installed in the folder that contained the .tar. If we call this folder <DYNAMO_ROOT>, you can activate Dynamo in your MATLAB session by typing:
>> run dynamo_activate.m
when you are in that folder, or with
>> run <DYNAMO_ROOT>/dynamo_activate.m
if you are in some other folder. Wait while some packages are compiled. Activate it again.
Amazon machines
If you want to follow the course with virtual machines provided by the course, follow this instructions.
Entering in the AWS Machine
Access the link of your AWS Machine provided by the organizers, that appears in the Google Sheet. After introducing your password (also given in the same google sheet), you will have entered in the AWS Machine.
Before accessing, it may be helpful to set the browser zoom to 50% or less and refresh the website. This will help later to visualize some GUIs in a friendlier way.
Activating Dynamo Standalone
Once you have accessed the AWS Machine, you need to activate Dynamo Standalone. Open the terminal emulator and type the next command:
source /home/student/dynamo-v-1.1.579/dynamo_activate_linux_shipped_MCR.sh
After some lines asserting the correct addition of several paths and tests, it should appear as the last line: "Dynamo is ready for standalone use in your Linux system".
Initializing Dynamo Standalone
For a specific explanation of the Standalone see [1]. Once dynamo has been initialized, you can enter in dynamo by typing
dynamo
You can also type
dynamo x
so dynamo will open a GUI console, instead of using the terminal, nevertheless the first option is strongly reccommended for some Walkthroughs.
Solution of some possible minor cons of working in a laptop =
Working with AWS Machine in the Work Shop may entails some minor cons that you may encounter. Here we present the most common ones so you can solve them quickly.
Problem opening a GUI Window
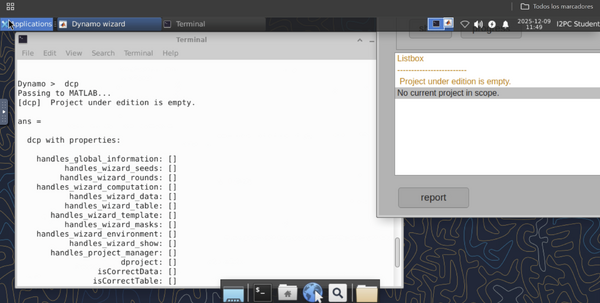
If you are working on a small laptop screen, opening a GUI window, for example when typing the "dcp" command to start a project, may make appear it with the header of the window hidden on the top of the screen:
A quick way of solving it is pressing Alt+Click on the window and move it down until the header appears so it is possible to change its size.
Problem pressing right click in a MAC laptop with no external mouse
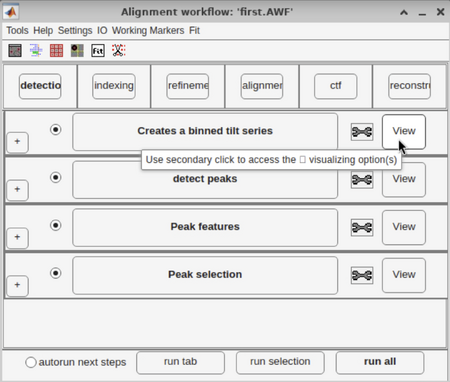
For some walkthroughs, like "Walkthrough on GUI based tilt series alignment", you need to do secondary click on the "View" botton:
In general, this corresponds to doing a right click, but if you are working on a MAC laptop with no external mouse, just press with two fingers on the trackpad at the same time.
Note that sometimes the GUI does not refresh itself and in this case, doing right click on the "View" botton will not work. To manually refresh the GUI and load all of the previous results into it, you need to do a primary click on the "View" bottom first. After that, you can do right click and you should be able to access the "View" panel as usual.
Unzipping some of the data
In the amazon machines, the data for the walkthrough on Geometry aware template matching is in a zipped file, so in order to access it, we need to unzip it. To do so, open a terminal and write:
cd /home/student/data/TemplateMatchingGUI
unzip MCP_walkthrough_data.zip
Workshop data
The data that will be used during the workshop can be downloaded through the SACO server of CSIC. In detail, the data for the respective modules is as follows (for a total of about 61 Gb ):
| Module | Data source |
|---|---|
| Starters Guide: subtomogram averaging | Data generated locally by Dynamo |
| Models: Filaments | Data generated locally by Dynamo |
| Models: Vesicles | Data generated locally by Dynamo |
| Models: Surfaces | Data generated locally by Dynamo |
| Template Matching | https://saco.csic.es/s/LAXyQgWnrLNFiym (750MB) |
| Template Matching (GUI) | https://saco.csic.es/s/SmymKiHxswSoyoP (7.6GB) |
| Alignment of tilt series | https://saco.csic.es/s/rWWRdGkYmbntywB (600MB) |
| Virus like particles | https://saco.csic.es/s/9mcg9wAZXqYe9FB (4.6GB) |
| Spherical geometries | https://saco.csic.es/s/smSJXzxNA2ynw8K (64 MB) |
| Principal Component Analysis | Data generated locally by Dynamo |
| Complete walkthrough on FHV virus | https://saco.csic.es/s/wmbDNPi2YcYFjKb (2GB) |
| Walkthrough on hexagonal lattice coherence through neighborhood analysis | https://saco.csic.es/s/T9EzcQ3qLxz9g4k |
| High-resolution HIV1 | https://saco.csic.es/s/JN5LGP4BbKSn9F8 (48Gb) |
Time schedule
| Module | Time | Subject |
|---|---|---|
| 1 | 09:30 - 10:30 | Hands-on tutorial. First steps and formats. |
| 2 | 10:30 - 11:00 | Starters Guide: subtomogram averaging |
| coffee | 11:00 - 11:30 | Coffee Break |
| 2 | 11:30 - 12:30 | Starters Guide: subtomogram averaging (continuation). |
| SBGrid | 12:30 - 13:00 | SBGrid Consortium presentation (Estrella Giménez) |
| 3 | 14:00 - 14:30 | General modelling |
| 3a | 14:30 - 15:00 | Models: Filaments Additional material: Coordinated extraction from several filaments |
| 3b | 15:00 - 15:15 | Models: Vesicles |
| 3c | 15:15 - 15:30 | Models: Surfaces |
| coffee | 15:30 - 16:00 | Coffee Break |
| 4 | 16:00 - 17:30 | Virus like particles or Spherical geometries |
The presentation introducing the automatic tilt series alignment, template matching, and principal component analysis walkthroughs can be found here.
| 5a | 09:30 - 09:45 | Presentation of FHV: Advanced starters guide |
| 5b | 09:45 - 11:00 | Hands-on tutorial: Advanced starters guide |
| coffee | 11:00 - 11:30 | Coffee break |
| 5b | 11:30 - 12:00 | Hands-on tutorial: Advanced starters guide (continued) |
| 6a | 12:00 - 12:20 | Alignment of tilt series: Manual |
| 6b | 12:20 - 12:40 | Alignment of tilt series: GUI |
| 6c | 12:40 - 13:00 | Alignment of tilt series: command line |
| Module | Time | Subject |
|---|---|---|
| 7a | 14:00 - 15:00 | Template Matching |
| 7b | 15:00 - 15:30 | Geometry aware template Matching |
| coffee | 15:30 - 16:00 | Coffee Break |
| 8 | 16:00- 17:00 | Principal Component Analysis |
Instructors
- Daniel Castano-Diez (CSIC Bilbao, Spain).
- Raffaele Coray (CSIC Bilbao, Spain).
- Laura García-Cornejo (CSIC Bilbao, Spain).
- Andrés Molina (CSIC Bilbao, Spain).
- Miguel Sánchez López-Cuervo (CSIC Bilbao, Spain).