Difference between revisions of "Viewing tomograms"
| Line 15: | Line 15: | ||
The main function of the <tt>dpreview</tt> is to provide a "preview" of the disk contents of a tomogram, letting the user to specify and area of the tomogram and a binning level. This fragment will be read into memory and passed to <tt>dtmview</tt> or <tt>dtslice</tt> | The main function of the <tt>dpreview</tt> is to provide a "preview" of the disk contents of a tomogram, letting the user to specify and area of the tomogram and a binning level. This fragment will be read into memory and passed to <tt>dtmview</tt> or <tt>dtslice</tt> | ||
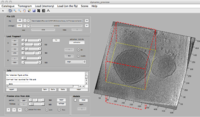
| − | [[File:dpreview_with_region.png]] | + | [[File:dpreview_with_region.png|200px| A screenshot on <tt>dpreview</tt> ]] |
===<tt>dtmview</tt> === | ===<tt>dtmview</tt> === | ||
Revision as of 15:22, 18 March 2016
Dynamo offers several ways to visualize tomograms. The choice of one or other program depends on varios factors, like "How big is the tomogram?" "Are you going to annotate models on them?".
Contents
Tomogram browsers
dtmshow
The simplest visualization system is the program dtmshow, callable from the command line and linked by many Dynamo GUIs. It's a lightweight slice-based browser. The user can choose to use it by preloading of the full tomogram or by accessing the disk for each depicted slice ("on-the-fly" modus). dtmshowDoes not allow to make complex annotations, or to register them inside the Catalogue via the model pool
dpreview
dpreview goes a step further that dtmshow. It is connected with the Catalogue system, in fact 'open with dpreview' is one of the main options that you get when selecting a catalogued tomogram in the dcm GUI.
In dpreviewslices are always accessed "on the fly", so that browsing can be slow (especially if trying to depict slices orthogonal to the x- or y- directions, which demand multiple I/O operations to gather a slice whose pixels are scattered in the disk). The main function of the dpreview is to provide a "preview" of the disk contents of a tomogram, letting the user to specify and area of the tomogram and a binning level. This fragment will be read into memory and passed to dtmview or dtslice
dtmview
Is normally invoked by the dcm GUI or by dpreview . It is oriented to show concurrently slices representing orthogonal cuts inside a volume with a high level of control of the number of depicted slices.
The main utility of dtmview is for clicking sets of isolated particles.
dtslice
Is normally invoked by the dcm GUI or by dpreview . It depicts in 3d a slide or arbitrary orientation that can be moved freely inside the tomogram. This is a suitable browser for filament geometries. It is also suitable to locate the extent of membranes a send them over to the Sliding Montage browser for annotation.
Sliding montage
Sliding montages of areas inside a tomogram are