Difference between revisions of "Walkthrough on GUI based tilt series alignment"
(→Data) |
|||
| Line 6: | Line 6: | ||
< tt> wget https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc</tt> | < tt> wget https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc</tt> | ||
| + | |||
| + | In a Mac: | ||
<tt>curl -O https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc</tt> | <tt>curl -O https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc</tt> | ||
| + | |||
| + | In the workshop, a copy should be already available on your local data folder: | ||
| + | |||
| + | <tt>~/data/b001ts001.mrc</tt> | ||
In this binned version, the pixel size is 2'7 Angstrom (original was 1.35, this is bin level 1 in <tt>Dynamo</tt>) | In this binned version, the pixel size is 2'7 Angstrom (original was 1.35, this is bin level 1 in <tt>Dynamo</tt>) | ||
| Line 22: | Line 28: | ||
The data for the workflow (tilt series, angles) could have been introduced directly when creating the workflow, we however will introduce each item explicitly in this walkthrough. | The data for the workflow (tilt series, angles) could have been introduced directly when creating the workflow, we however will introduce each item explicitly in this walkthrough. | ||
| − | |||
The minimal data to initiate a workflow is | The minimal data to initiate a workflow is | ||
| − | * tilt series matrix (use the | + | * tilt series matrix (use the file indicated above) |
* tilt angles. | * tilt angles. | ||
* discarded tilt angles (leave empty) | * discarded tilt angles (leave empty) | ||
| Line 79: | Line 84: | ||
=== Detection of gold beads === | === Detection of gold beads === | ||
| − | + | This area is the one that needed some design decision on the size of the gold bead. We will use <tt>dtmshow</tt> | |
[[ File:alignGUI_completedStepsBecomeBoldFontSize.png |thumb|center| 600px|completed steps become bold font size]] | [[ File:alignGUI_completedStepsBecomeBoldFontSize.png |thumb|center| 600px|completed steps become bold font size]] | ||
| − | |||
[[ File:alignGUI_thePlusIconAtCornerShowsTheValuesOfTheParametersOfAGivenStep.png |thumb|center| 600px|the plus icon at corner shows the values of the parameters of a given step]] | [[ File:alignGUI_thePlusIconAtCornerShowsTheValuesOfTheParametersOfAGivenStep.png |thumb|center| 600px|the plus icon at corner shows the values of the parameters of a given step]] | ||
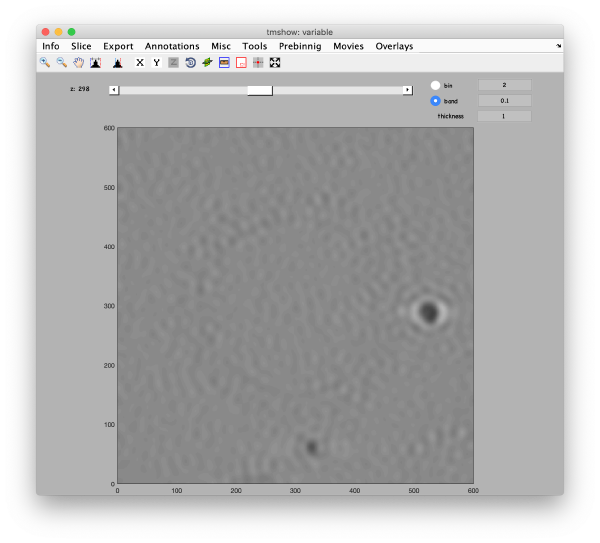
[[ File:alignGUI_openTomoshowOnTheFullSizedMatrixToCheckTheSizeOfAGoldBead.png |thumb|center| 600px|open tomoshow on the full sized matrix to check the size of a gold bead]] | [[ File:alignGUI_openTomoshowOnTheFullSizedMatrixToCheckTheSizeOfAGoldBead.png |thumb|center| 600px|open tomoshow on the full sized matrix to check the size of a gold bead]] | ||
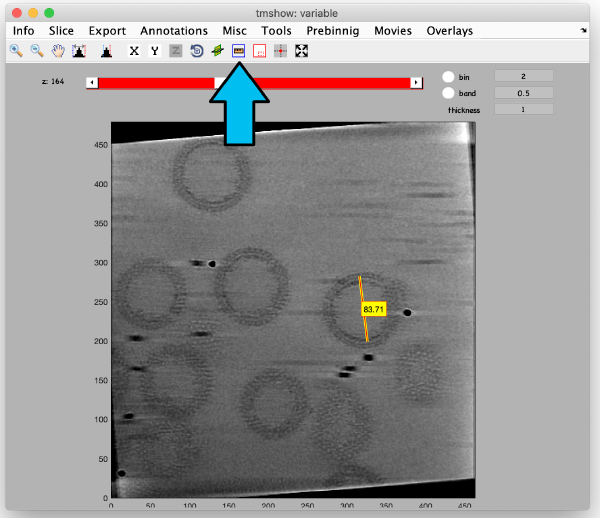
[[ File:alignGUI_theRulerToolAllowsToMeasureInPixelsTheDiameterOfAGoldBead.png |thumb|center| 600px|the ruler tool allows to measure in pixels the diameter of a gold bead]] | [[ File:alignGUI_theRulerToolAllowsToMeasureInPixelsTheDiameterOfAGoldBead.png |thumb|center| 600px|the ruler tool allows to measure in pixels the diameter of a gold bead]] | ||
| + | |||
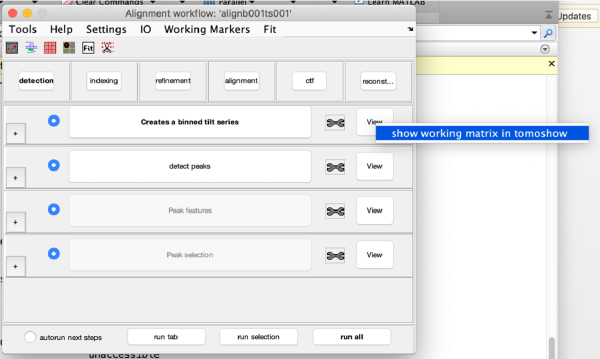
| + | Right click on the <tt>view</tt> area of the step to access the different visualisation options. | ||
[[ File:alignGUI_secondaryClickInTheViewPanelOpensAMenuToAccessTheDifferentResultsOfAStep.png |thumb|center| 600px|secondary click in the view panel opens a menu to access the different results of a step ]] | [[ File:alignGUI_secondaryClickInTheViewPanelOpensAMenuToAccessTheDifferentResultsOfAStep.png |thumb|center| 600px|secondary click in the view panel opens a menu to access the different results of a step ]] | ||
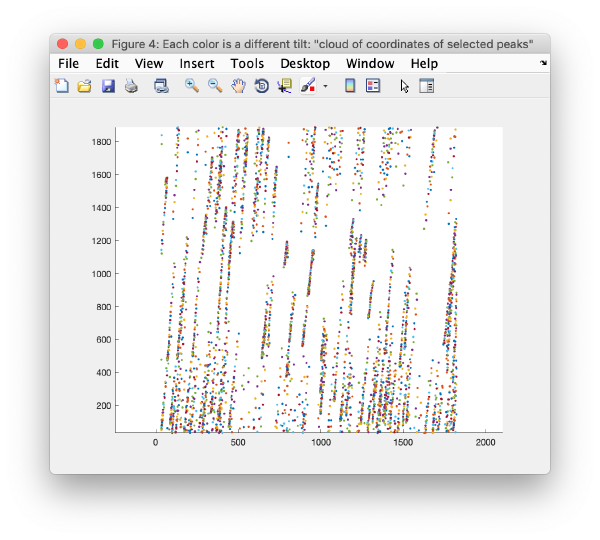
[[ File:alignGUI_markerSetsCanBeViewedAsCloudOfPoints.png |thumb|center| 600px|marker sets can be viewed as cloud of points]] | [[ File:alignGUI_markerSetsCanBeViewedAsCloudOfPoints.png |thumb|center| 600px|marker sets can be viewed as cloud of points]] | ||
| + | |||
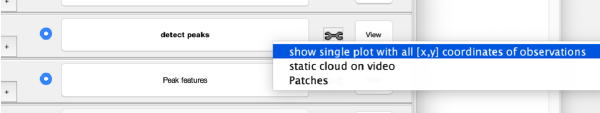
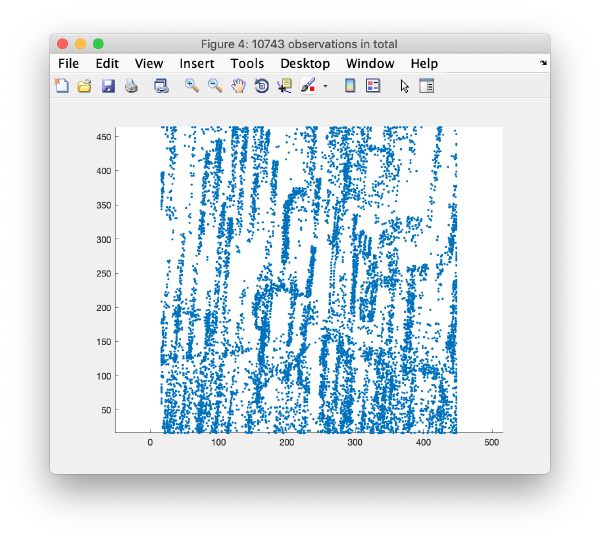
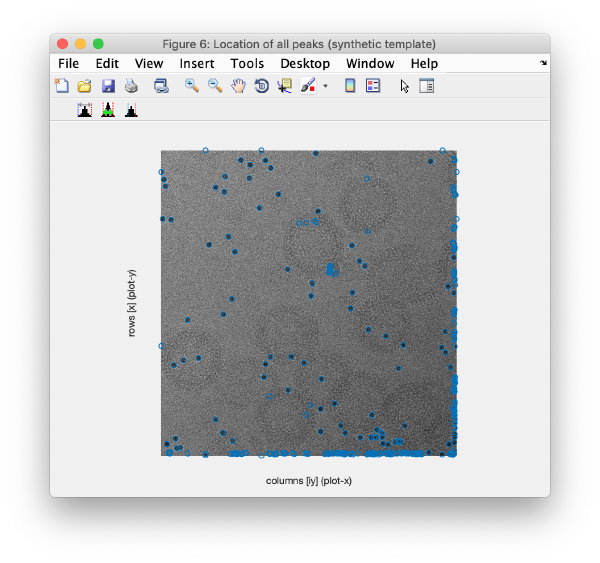
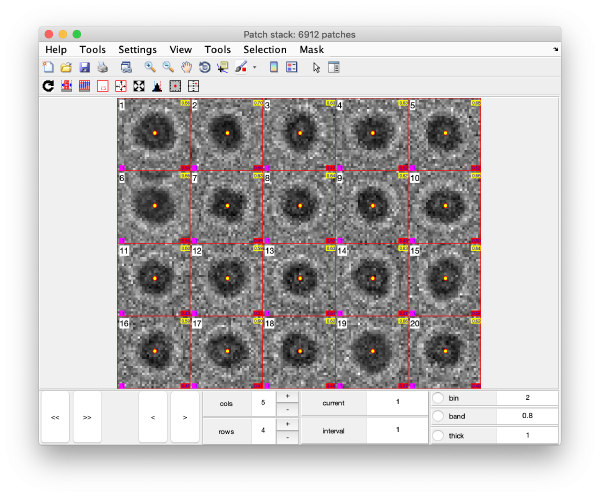
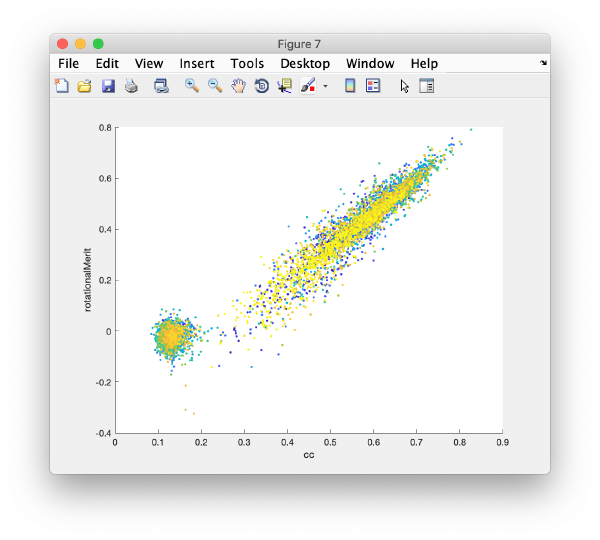
| + | This view shows the x,y coordinates of ''all'' cross correlation peaks on | ||
[[ File:alignGUI_cloudOfUnindexedCcPeaks.png |thumb|center| 600px|cloud of unindexed cc peaks]] | [[ File:alignGUI_cloudOfUnindexedCcPeaks.png |thumb|center| 600px|cloud of unindexed cc peaks]] | ||
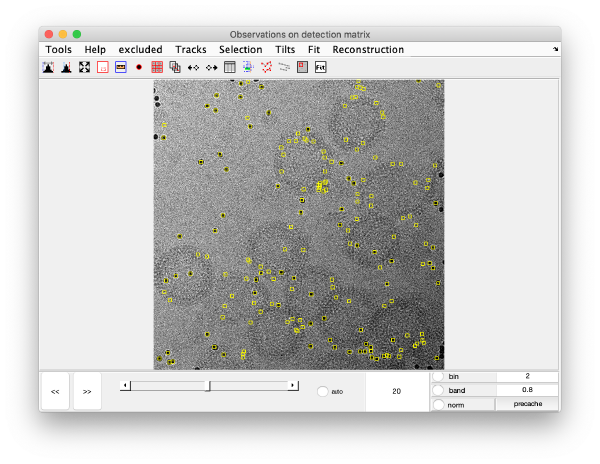
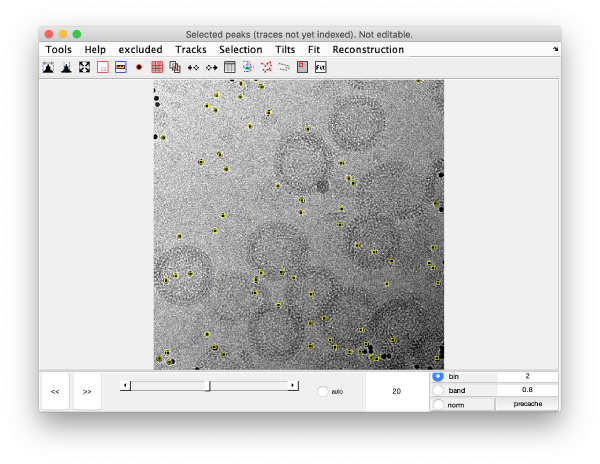
[[ File:alignGUI_showsTheDetectedSpotsOnTheVideoGUI.png |thumb|center| 600px|shows the detected spots on the video GUI]] | [[ File:alignGUI_showsTheDetectedSpotsOnTheVideoGUI.png |thumb|center| 600px|shows the detected spots on the video GUI]] | ||
Revision as of 14:57, 27 August 2019
Dynamo includes a package for automated aligned and reconstruction of tilt series. This walkthrough guides you through the steps on how to use it with a GUI. A different page in this wiki
Contents
Data
We have prepared a version of a tilt series from the EMPIAR entry [XXXX], containing a set of virus like particles of VLP.
< tt> wget https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc
In a Mac:
curl -O https://wiki.dynamo.biozentrum.unibas.ch/html/w/doc/data/hiv/align/b001ts001.mrc
In the workshop, a copy should be already available on your local data folder:
~/data/b001ts001.mrc
In this binned version, the pixel size is 2'7 Angstrom (original was 1.35, this is bin level 1 in Dynamo)
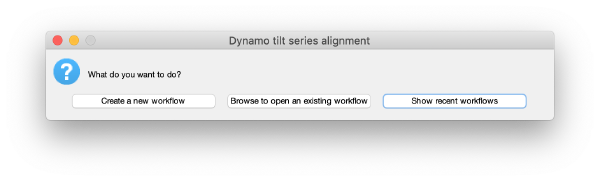
Creating an alignment workflow
The basic function for invoking the GUI is dtsa (short form of dynamo_tilt_series_alignment;
u = dtsa();
Here, u is an object that will reside in memory during the session and allows to interoperate with the workflow through the command line. It is not necessary when proceeding through the GUI.
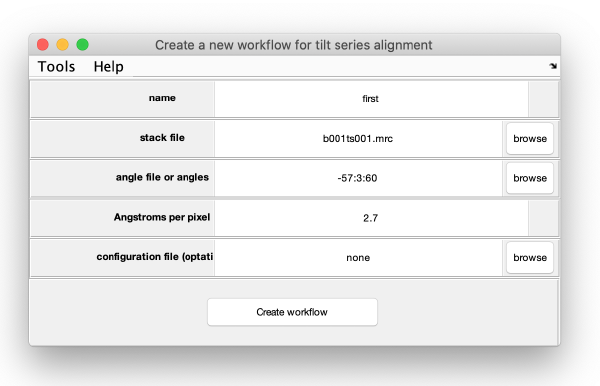
The data for the workflow (tilt series, angles) could have been introduced directly when creating the workflow, we however will introduce each item explicitly in this walkthrough.
The minimal data to initiate a workflow is
- tilt series matrix (use the file indicated above)
- tilt angles.
- discarded tilt angles (leave empty)
Discarded tilt angles
If you are unsure on the tilts to be discarded, you can do it in a later stage while you inspect the images in the dmarkers GUI, using the x and shift + x buttons.
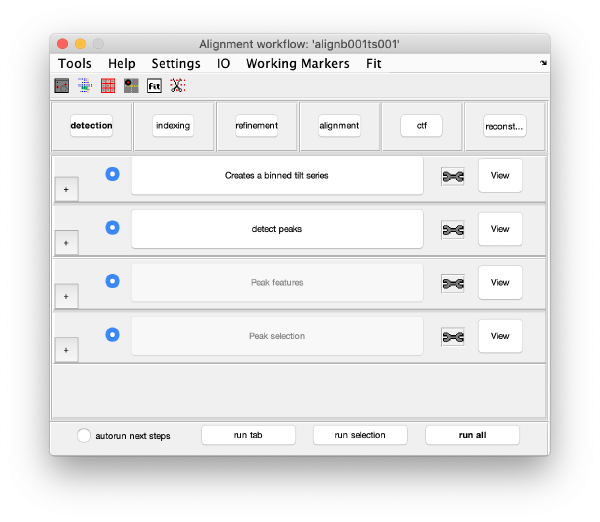
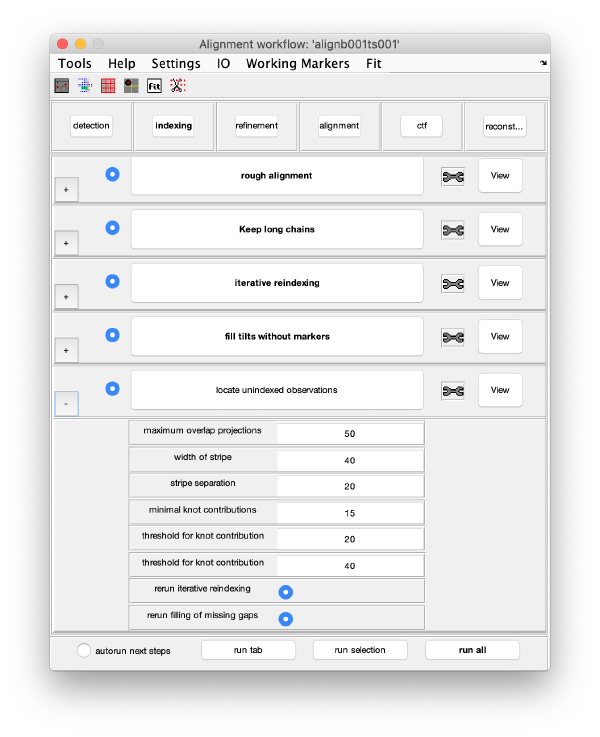
GUI description
The GUI guides of the process of creating and editing a set of "working markers" (in the areas of detection, reindexing and refinement), and then using them to create aligned stacks (alignment tab), analyse and correct their CTF (through wrappers to ctfffind4 and imod) and then create reconstructions.
Settings
Acquisition settings
Here you can enter pixel size (2.7 Angstroms), Cs (2.2), nominal defocus (-4.0 microns, used if the CTF area is connected).
Controls
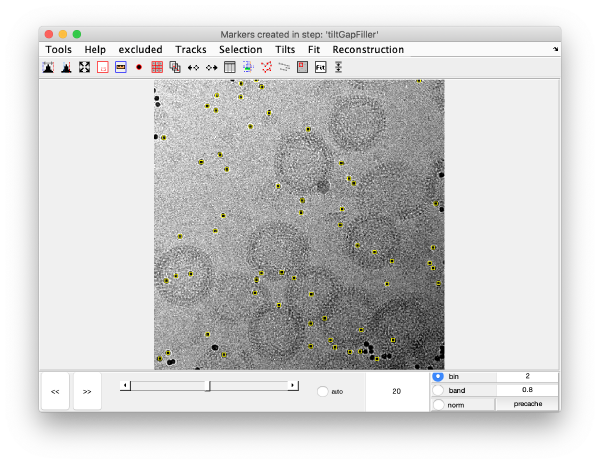
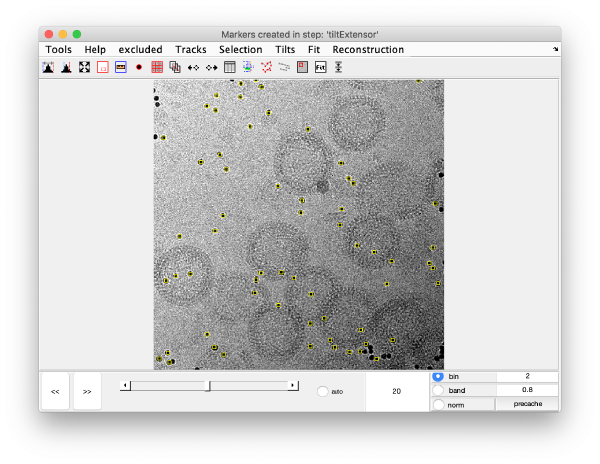
markers GUI
The icon of the markers GUI allows you to check the position of the working markers in the micrograph. If you want to make editions of the GUI permanent, you need to save the current markers.
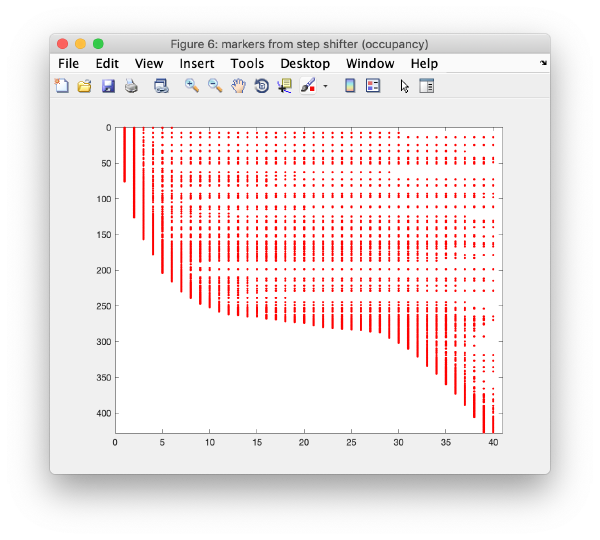
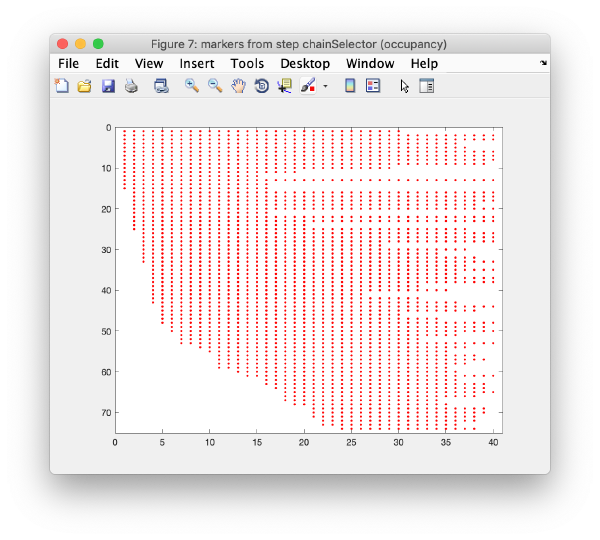
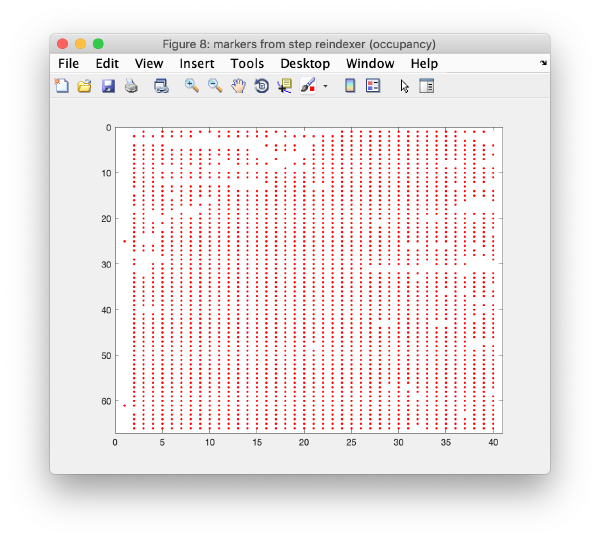
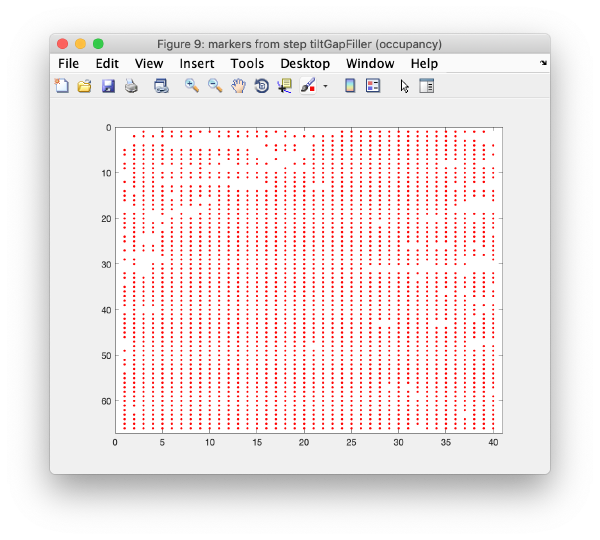
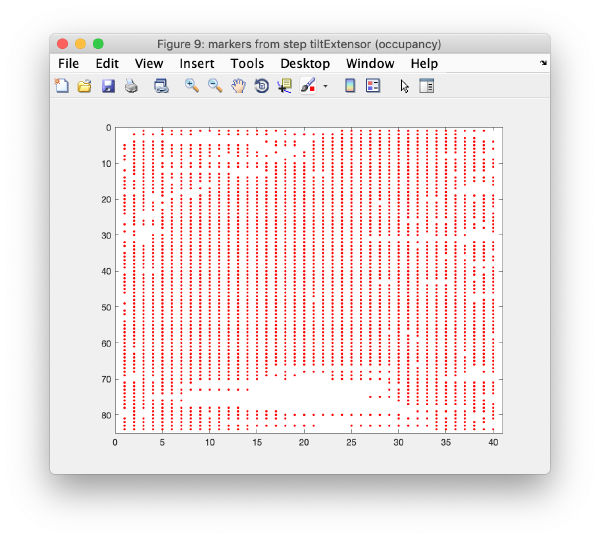
Occupancy plot
Shows a scheme of which markers are present in which tilt numbers.
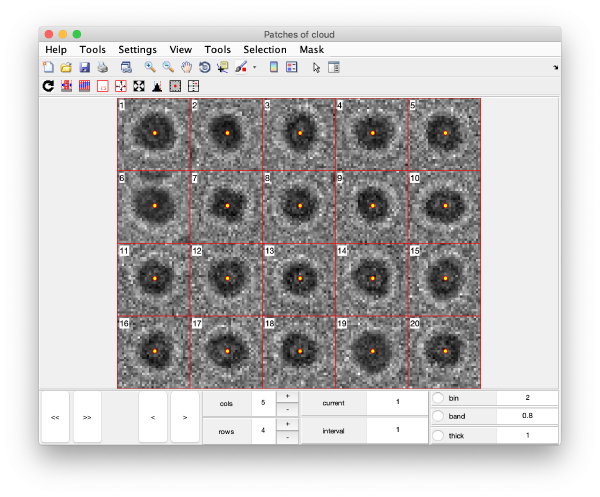
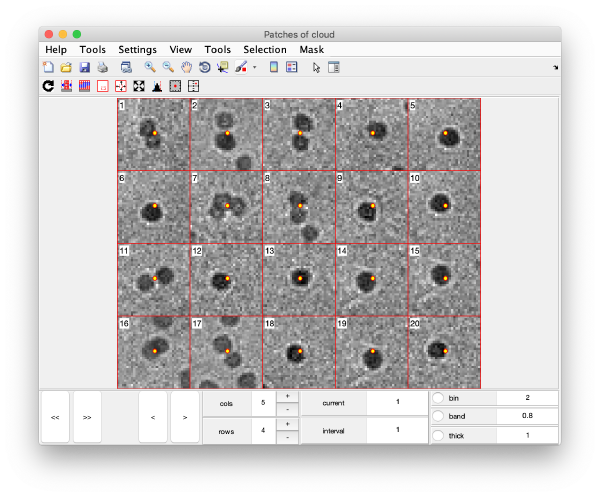
Gold bead gallery
Crops the gold beads currently contained in the working markers and shows them as a gallery.
Execution Areas
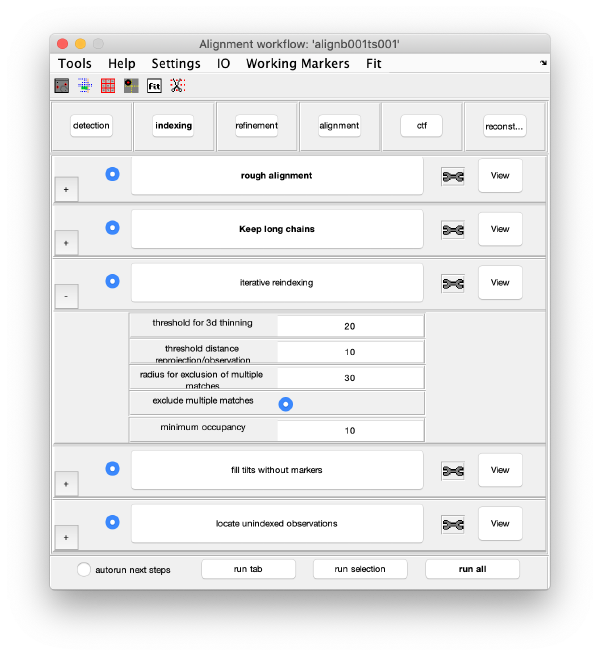
The GUI is divided in five areas. Each are comprises sets of steps that can be executed sequentially. Sequential execution is the natural way of proceeding; but steps can be visited in any order, i.e., a step can be reexecuted in a later moment after updating its information.
Steps
Steps needs to be open (letters in black, not grey) in order to be executed. They become open when the input they require gets computed and stored by a previous step during execution of the workflow. When a step is under execution, its pushbutton becomes green.
Step Toolbox
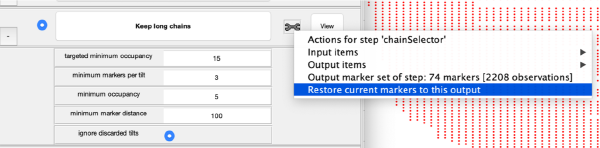
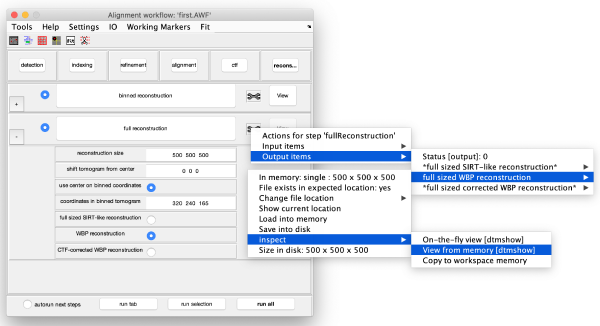
The tool icon can be secondary clicked and will provide a popup menu with specific tools for each steps and some common one. The common ones are handles to inspect and list the items that are needed (input items) or generated (output items) by the step. The View provides tools specific for visualisation of results of the step.
The areas in the alignment task
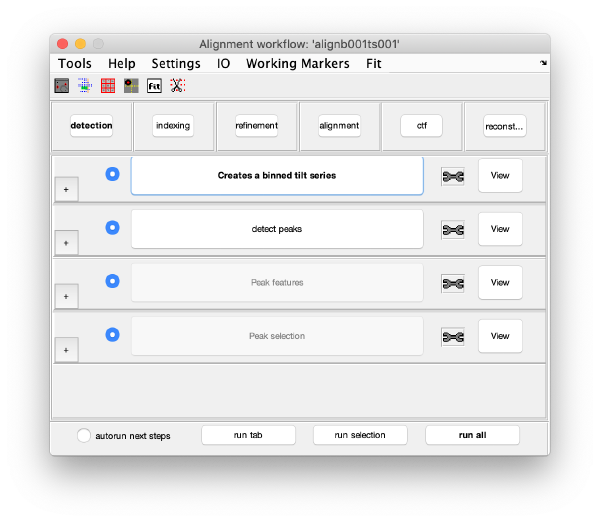
Detection
The goal of this task is to find the positions of the gold bead projections in the tilt series.
Visualization matrix
This step just creates a binned version of the tilt series stack to accelerate visualizations that will be invoked in a later point.
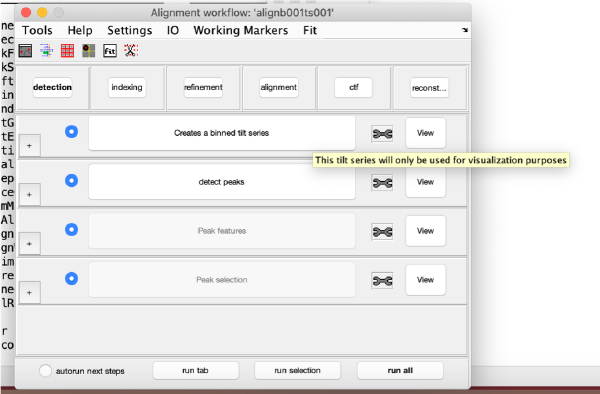
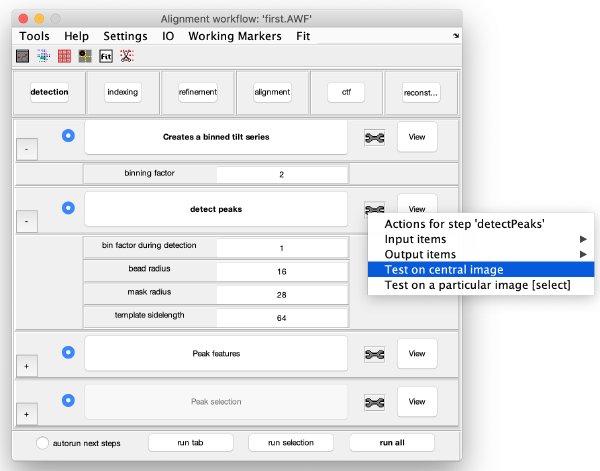
Detection of gold beads
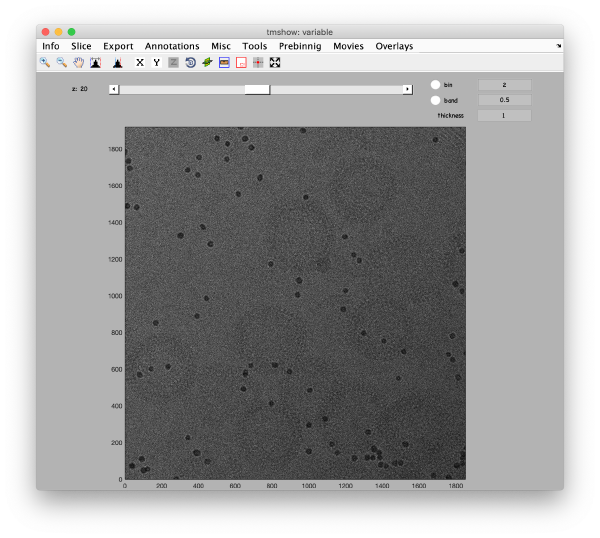
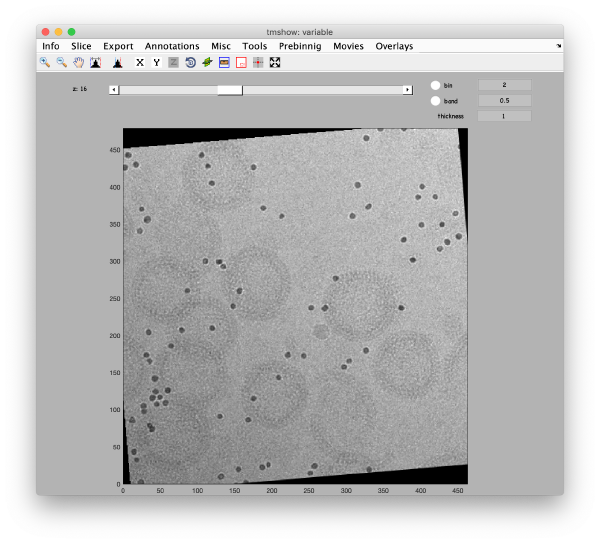
This area is the one that needed some design decision on the size of the gold bead. We will use dtmshow
Right click on the view area of the step to access the different visualisation options.
This view shows the x,y coordinates of all cross correlation peaks on
Computation of observation features s
Selection of best gold beads
Indexing
The goal of this task is to find trails of gold beads, i.e., observations in different micrographs that correspond to the same 3d gold bead.
Rough Alignment
Here all couples of micrographs are compared.
Selection of stable trails
Iterative reindexing
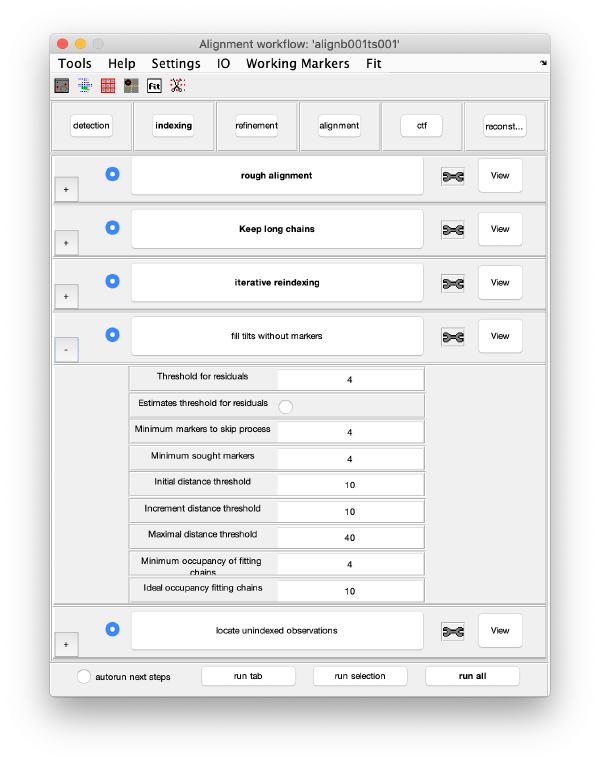
Tilt gap filling
Trail extension
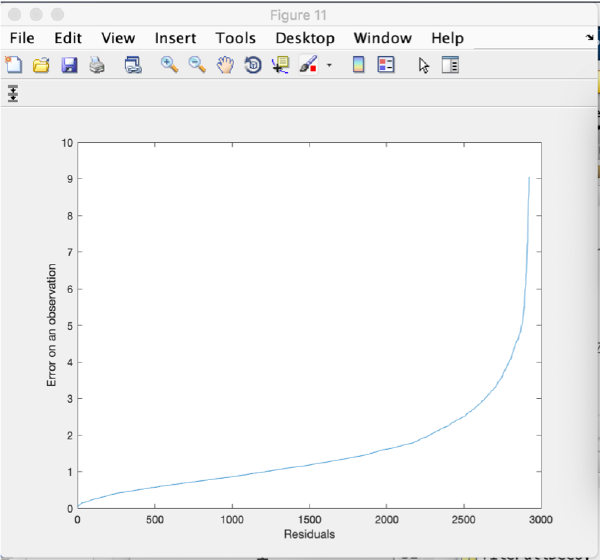
Refinement
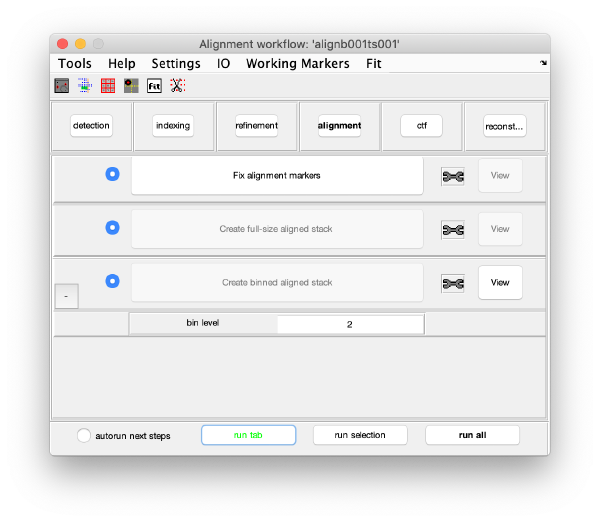
Alignment
Fix the markers
Align the tilt series
CTF
In this GUI CTF estimation and correction are delegated to CTFFIND4 and Imod's ctfphaseflip programs respectively. This part will not be used in the workshop.'
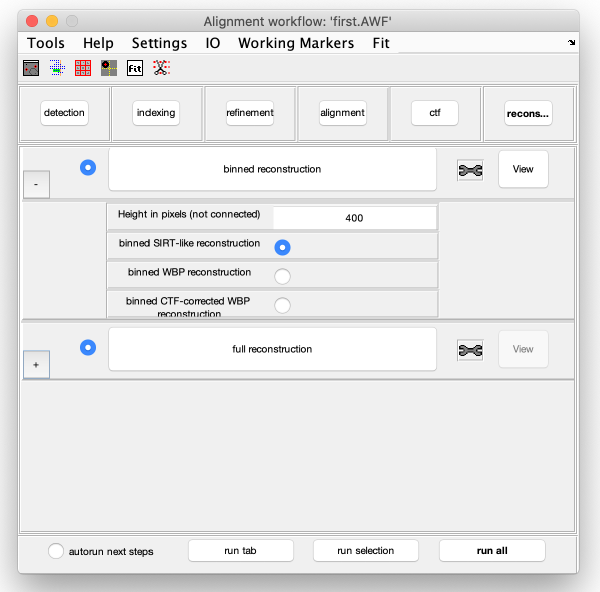
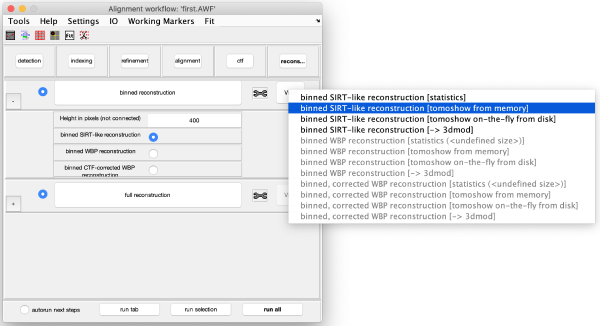
Reconstruction
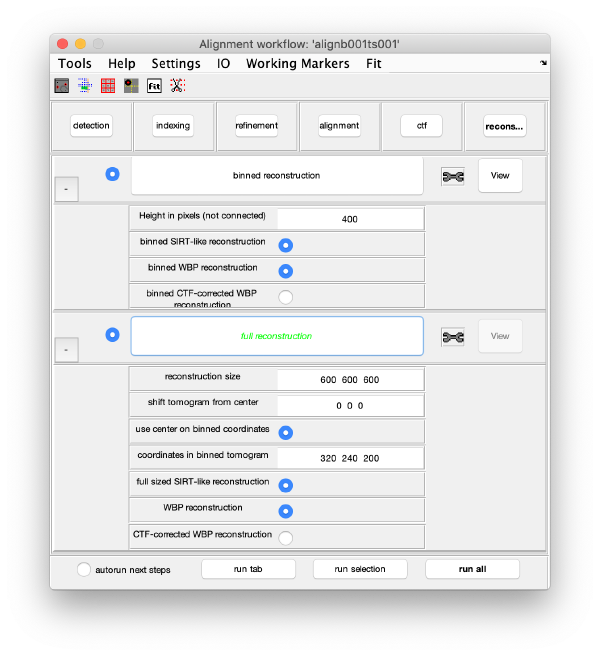
Binned reconstruction
Full sized reconstruction
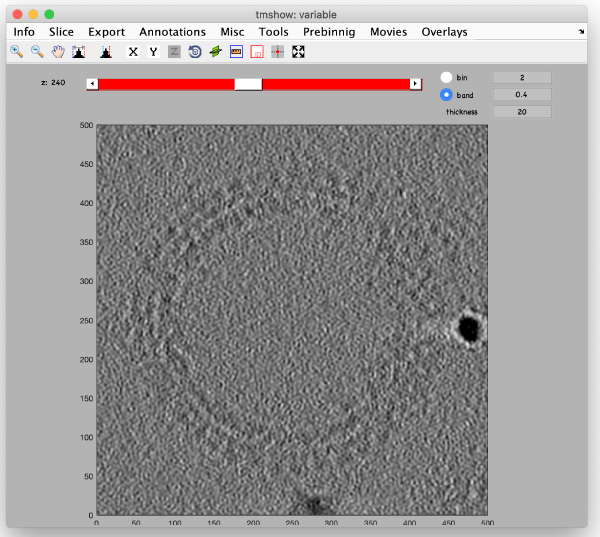
The full sized reconstruction can be created related to a coordinate directly read in the binned tomogram, a feature useful for creating reconstructions localised on an area of interest. In this case, we will focus on one of the VLPS.
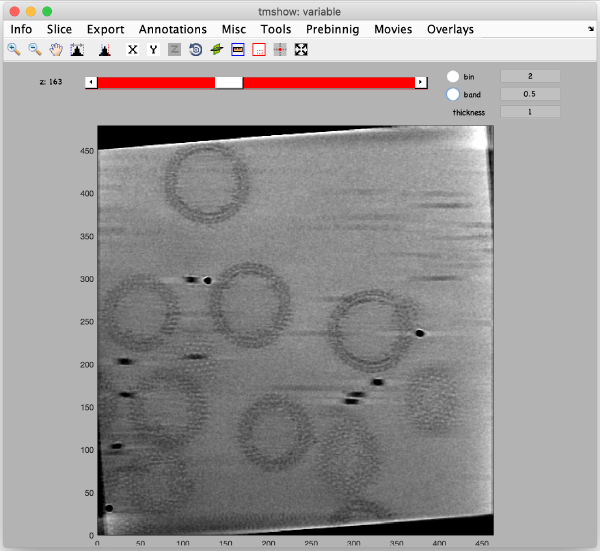
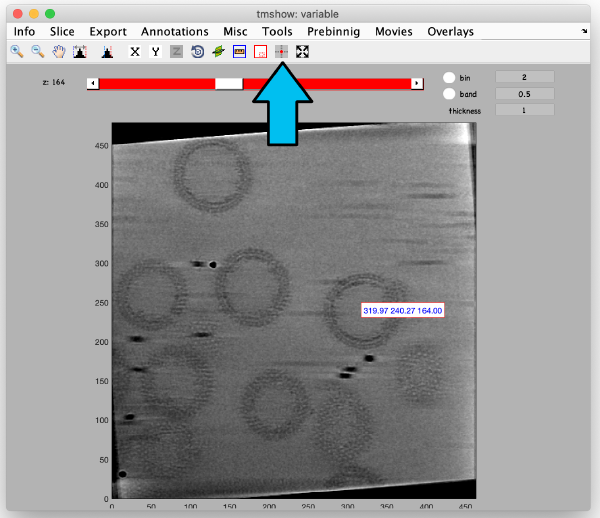
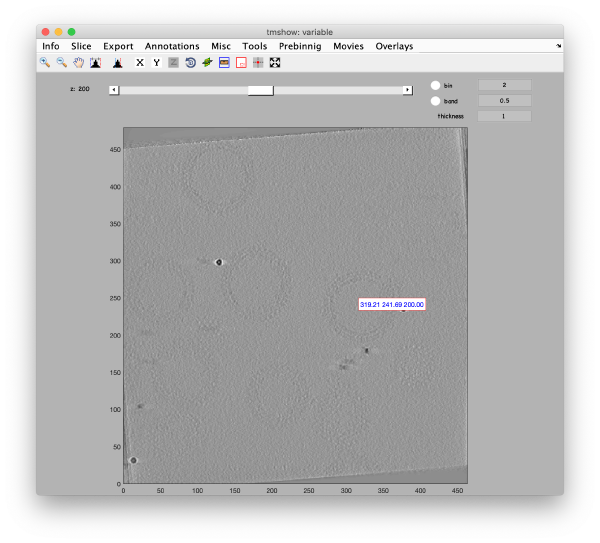
Use the pin tool to select a point on the binned reconstruction.
We also check the size of a structure of interest. We measure it in the binned tomogram.
]