EMBO workshop 2023
The tutorials listed here take place virtually on the 9th and 10th of September of 2023 (13:30-18:00, London time) as part of the EMBO workshop for Image processing in cryoEM.
Contents
Connect to Guacamole and run Dynamo
Credentials
Please refer to your personal course material to access your two sets of credentials:
- A set of credentials to enter Guacamole (the platform for connecting via browser). These are common to all participants.
- A set of course credentials to enter the actual machines on which the course will be done: the EMBO21 UserID. These are unique to each participant.
Connect to Guacamole
To connect to Guacamole, first go to the following link in your browser:
https://login.cryst.bbk.ac.uk/guacamole/#/
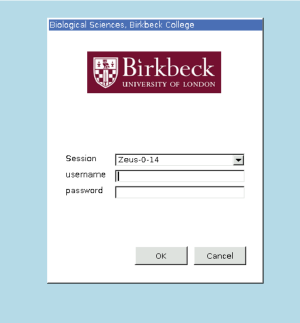
At this stage you should maximize your browser window, otherwise you might have visualization problems when opening Dynamo. You should see the following window. Enter the credentials of the workshop (common to all participants; do not use the ones that are particular to you.) .
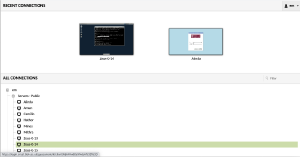
Connect to your machine
Now you need to select a machine. Please then click on +em and then +Servers - Public and select your dedicated server (in this example it is Zeus-0-14).
When you select the machine, you will prompted to use your EMBO23 UserID
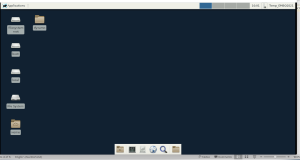
This should access your virtual Linux XFCe desktop, which should look like this in your browser
If you open a terminal (with the terminal icon) you can test by typing
pwd
that you are in the correct path, which should be:
/d/embo2021/u/emboXX
where XX stands for your actual workshop participant number.
Data location
The data used in the tutorials is already available for you. Move to the directory of the corresponding practical using the command:
cd prac-5
or
cd prac-6
respectively. Double-check if the data is present by typing
ls
You should see the following files in both directories:
During the Dynamo workshop, work only within these two directories.
Load and run Dynamo from Matlab
To use Dynamo, you first need to open Matlab. For this, write in a terminal:
matlab
Then, inside Matlab type:
run /s/ems/s/dynamo/v1.1.541/dynamo_activate.m
Chimera
To use chimera, use the following command (replace 'myFile.mrc' with your file) in the normal terminal (outside Dynamo).
/usr/local/bin/chimera myFile.mrc
Schedule
The course is divided into short presentations and tutorials, as described in the time schedule below. For the presentations, we all meet in the main room. The tutorials are structured as follows:
- Demonstration: All students stay connected to the main room for the first minutes, where the goals of the tutorial are briefly explained.
- Individual work: Students go the assigned breakout rooms to work on the material independently. An instructor will be assigned to each breakout room to help with questions.
- Closure: During the last 5 minutes we come back to the general breakout room to wrap up the tutorial.
| Day 1 | |||
|---|---|---|---|
| Time (London) | Activity | Topic | Comment |
| 13:30 - 13:45 | Presentation | Introduction | |
| 13:45 - 14:00 | Tutorial | Alignment of tilt series: Manual | |
| 14:00 - 14:30 | Tutorial | Tilt series alignment with GUI | |
| 14:30 - 15:00 | Tutorial | Tilt series alignment with command line | |
| 15:00 - 15:15 | Break | ||
| 15:15 - 15:45 | Tutorial | Models: Filaments Additional material: Coordinated extraction from several filaments |
|
| 15:45 - 16:15 | Tutorial | Models: Vesicles | |
| 16:15 - 17:00 | Tutorial | Models for surfaces | |
| 17:00 - 18:00 | Tutorial | Template matching | |
| Day 2 | |||
| Time (London) | Activity | Topic | Comment |
| 13:30 - 13:45 | Presentation | Introduction | |
| 13:45 - 15:45 | Tutorial | Starters guide | |
| 15:45 - 16:30 | Tutorial | PCA | |
| 16:30 - 17:55 | Tutorial | Advanced starters guide | Take a 15 min break individually during this tutorial |
| 17:55 - 18:00 | Presentation | Closing remarks | |
| Optional: 18:00 | Tutorials | Virus like particles or Spherical geometries | |